Features
MiMiC currently supports using CPMD as the main MD driver and for calculating subsystem contributions it has interfaces to CPMD and GROMACS. Using the currently supported programs it is possible to run electrostatic-embedding QM/MM MD simulations where CPMD can be used as the QM engine and GROMACS as the MM engine. MiMiC also supports using the PLUMED library for performing enhanced-sampling QM/MM MD simulations.
Parallel Scalability
The MiMiC-based QM/MM implementation has displayed strong scalability well beyond ten thousand cores in a single QM/MM simulation while maintaining an overall parallel efficiency of at least 70%. 2
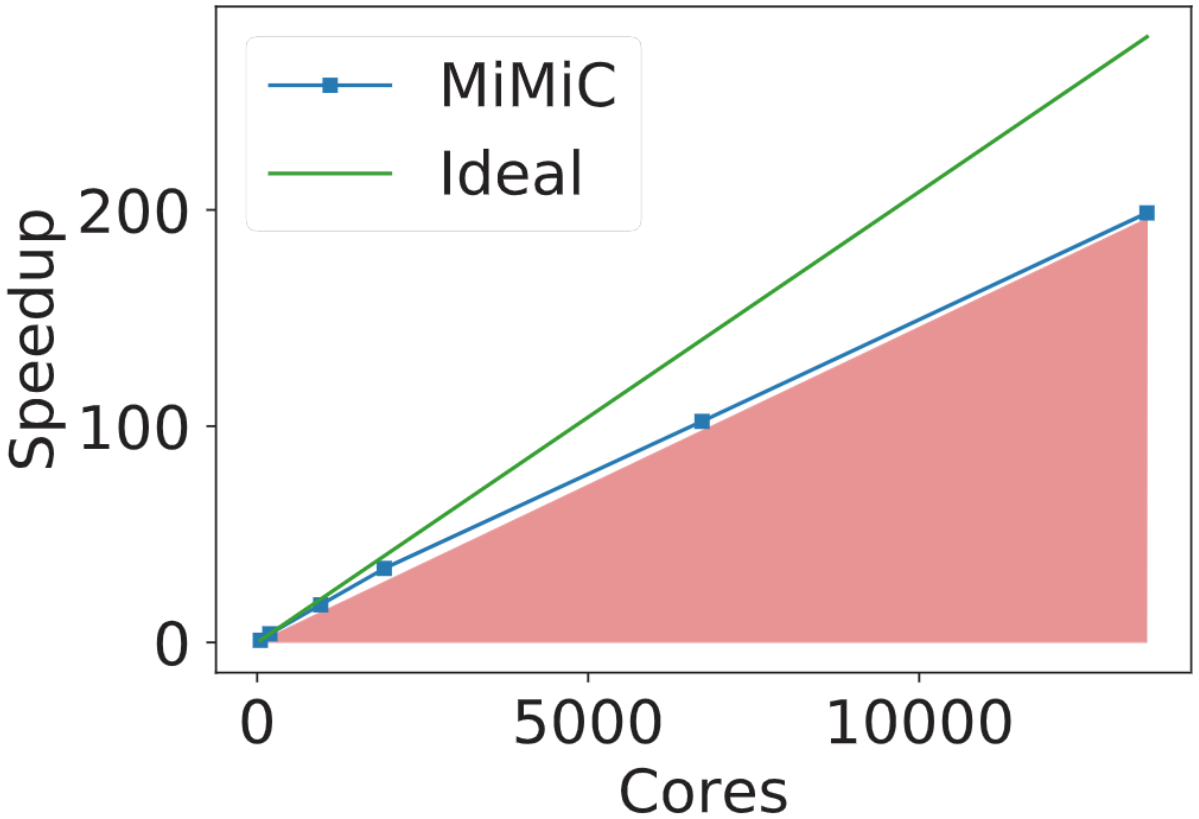
Scaling performance within Born-Oppenheimer molecular dynamics for a system containing a large Cl−/H+ antiporter protein embedded in a lipid membrane bilayer solvated in water. In this system, 19 atoms out of a total of 150,925 atoms were treated at the B3LYP QM level. Simulations were run on the JUWELS cluster at the Juelich Supercomputing Center.
Applications
MiMiC-based QM/MM MD simulations combined with metadynamics have been successfully applied to investigate complex biochemical processes. Below are some examples.
Molecular basis of CLC antiporter inhibition by fluoride
CLC channels and transporters conduct or transport various kinds of anions, with the exception of fluoride, which acts as an effective inhibitor. This study identified the high affinity of both F− and E148 for protons as the basis of the transport inhibition of the CLC anion/proton exchangers from E. coli. 3
Sub-nanosecond QM/MM simulations of the E. coli anion/proton exchanger ClC-ec1 showed that fluoride binds incoming protons within the selectivity filter, with excess protons shared with the gating glutamate E148. Depending on E148 conformation, the competition for the proton can involve either a direct F−/E148 interaction or the modulation of water molecules bridging the two anions. The direct interaction locks E148 in a conformation that does not allow for proton transport, and thus inhibits protein function.
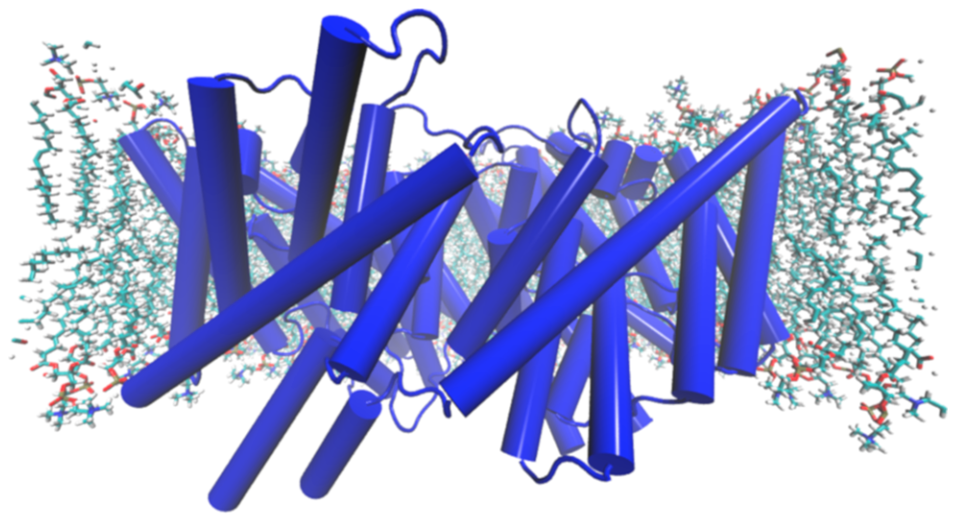
E. coli CLC antiporter (CLC-ec1) in a lipid bilayer in the simulation setup. Water and counterions are not shown for clarity.
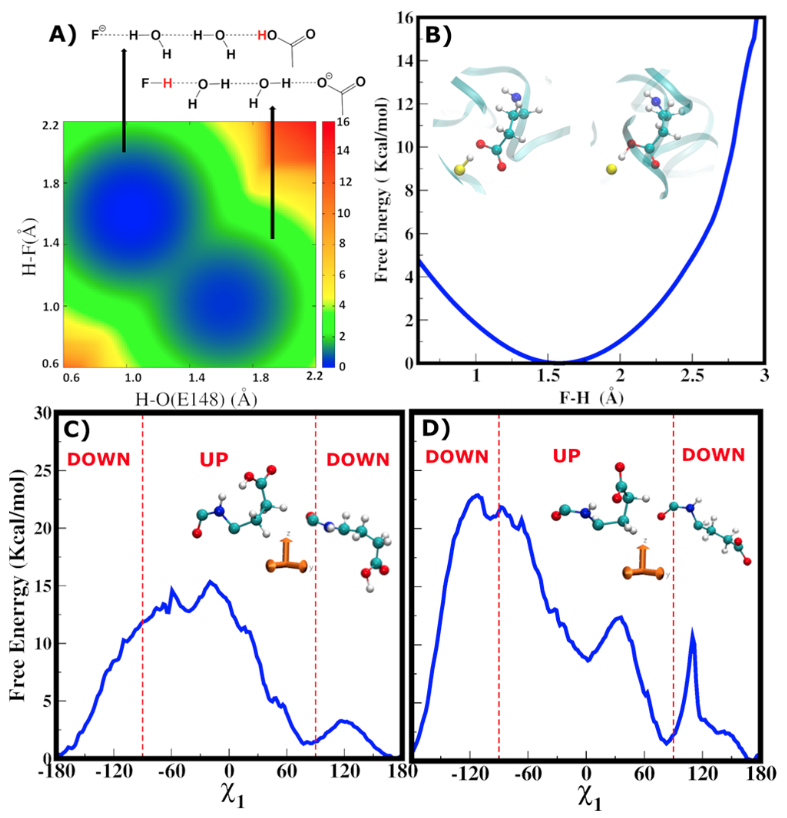
Free energy profiles (in kcal/mol) emerging from QM/MM metadynamics simulations. (A) Free energy associated with the proton transfer (PT) process between fluoride and E148 modulated by two water molecules. The schematic representation of the two free energy minima is displayed on the top of the figure. Here, E148 is in the up conformation. (B) Free energy profile associated with the direct PT process between F− and E148 in the down conformation. The minimum is reached at H−F and H−O (E148) distances of 1.5 and 1.0 Å, respectively. No barrier separates the state in which the proton is bound to the fluoride (H−F 1.0 Å). (C, D) Free energy as a function of E148’s \(\Chi_1\) dihedral angle for E148 protonated (C) and deprotonated (D). The relative positions of the E148 carboxyl group above or below the backbone unit define the up or down conformations. The transition from one conformation to the other is indicated by dashed red lines.
Mechanisms underlying proton release in CLC-type F−/H+ antiporters
The CLC family of anion channels and transporters includes Cl−/H+ exchangers (blocked by F−) and F−/H+ exchangers (or CLCFs). CLCFs contain a glutamate (E318) in the central anion-binding site that is absent in CLC Cl−/H+ exchangers. The X-ray structure of the protein from Enterococcus casseliflavus (CLCF-eca) shows that E318 tightly binds to F− when the gating glutamate (E118; highly conserved in the CLC family) faces the extracellular medium. This study illustrated how glutamate insertion into the central anion-binding site of CLCF-eca permits the release of H+ to the cytosol as HF, thus enabling a net 1:1 F−/H+ stoichiometry. 4
Classical and MiMiC-based QM/MM metadynamics simulations were used to investigate proton transfer and release by CLCF-eca. Results show that after up to down movement of protonated E118, both glutamates combine with F− to form a triad, from which protons and F− anions are released as HF.
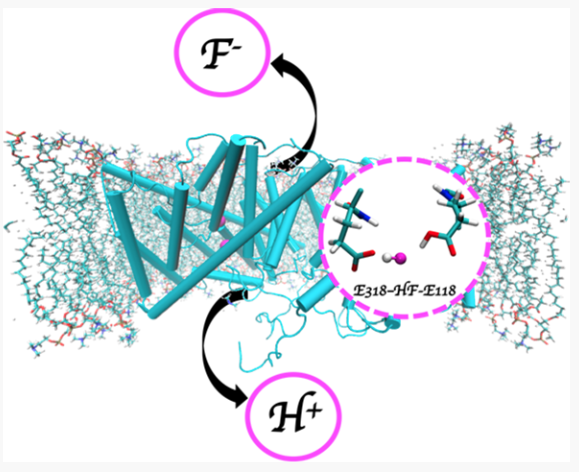
E. casseliflavus F−/H+ antiporter (CLCF-eca) embedded in a lipid bilayer in the simulation setup. Water and counterions are not shown for clarity. The inset show HF interacting with the two glutamates E118 and E338.
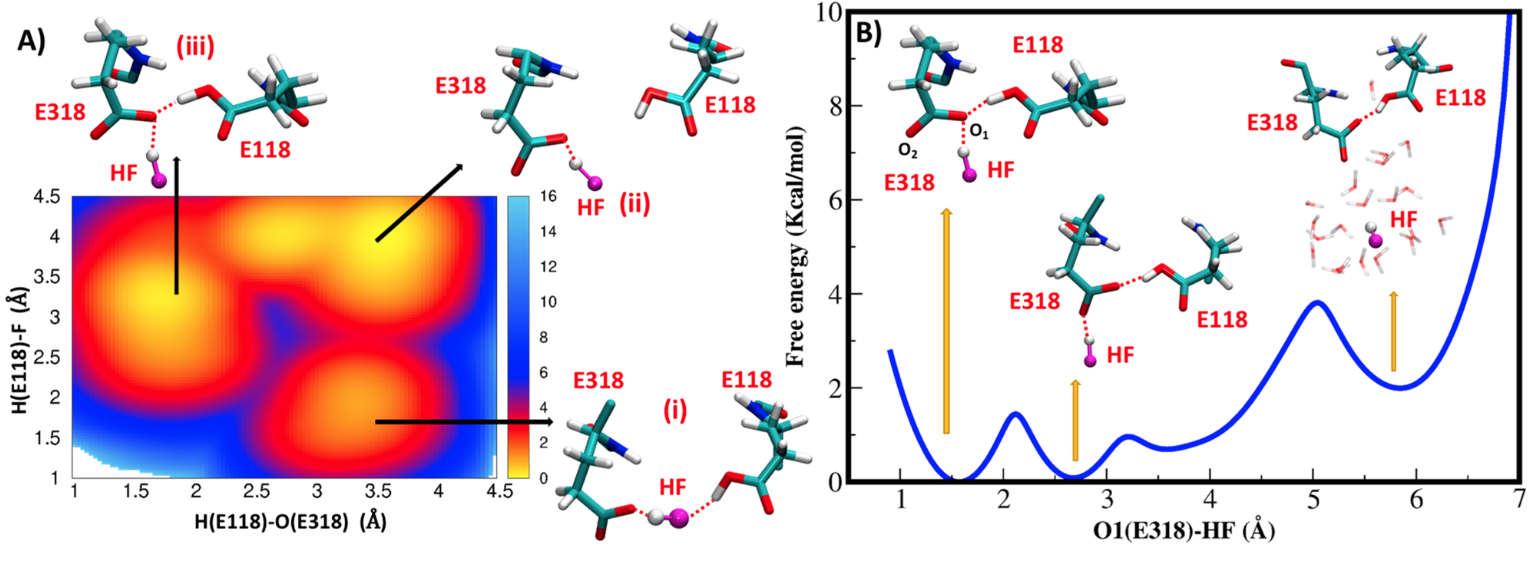
(A) Free energy landscape of conversion of the E318−F−E118 triad into the intermediate conformation with E318 and E118 in direct contact. (B) Free energy profile of HF release into the intracellular solution. The E318 carboxyl oxygen atoms are labeled O1 and O2. H-bonds are shown as red dashed lines and free energies are in kcal/mol.